import numpy as np
from scipy.stats import poisson
from scipy.stats import expon
import matplotlib.pyplot as plt
from IPython.display import Image, display
import seaborn as sns
# Apply the default theme
sns.set_theme()
Exercise#
Exercise 1 - Frequentist Upper Limit on a Poisson Parameter#
Consider the scenario where the observed count n follows a Poisson distribution:
Given that the background expectation is b = 4.5 and the observed count is \(n_{\text{obs}} = 5\) , determine the 95% confidence level (CL) upper limit on s.
Hint
How does the distribution of n change as you vary s?
Exercise 2 - Maximum Likelihood Estimator for a Gaussian Distribution#
Suppose individual measurements \(x_i\) are distributed around an unknown true value \(\theta\) according to a Gaussian distribution with a known standard deviation \(\sigma_i\). The probability density function (PDF) is given by:
The corresponding log-likelihood function is:
This function describes a parabola, which reaches its maximum at some value \(\hat{\theta}\). Derive the expressions for the maximum likelihood estimator \(\hat{\theta}\), and its uncertainty \(\sigma_{\hat{\theta}}\).
Exercise 3 - Parameter Estimation for Exponential Decay#
The probability density function (PDF) for observing a decay at time \(t\) is given by:
The goal is to estimate the parameter \(\tau\), which represents the lifetime of the decay. Three different datasets, each containing a different number of events, are provided. Using the maximum likelihood estimation (MLE) method, calculate \(\hat{\tau}\) and \(\sigma_{\hat{\tau}}\) for each dataset and compare the results with those obtained using the Gaussian estimator.
Note
For any probability density function \(f(x;\theta)\), as the number of observed events increases, the likelihood function \(L\) asymptotically approaches a multivariate Gaussian distribution.
# set the random seed
np.random.seed(0)
Ns = [2, 8, 20]
true_tau = 1
# generate the data
toys_expon = [expon.rvs(size=N, scale=true_tau) for N in Ns]
Exercise 4 - Neyman Construction#
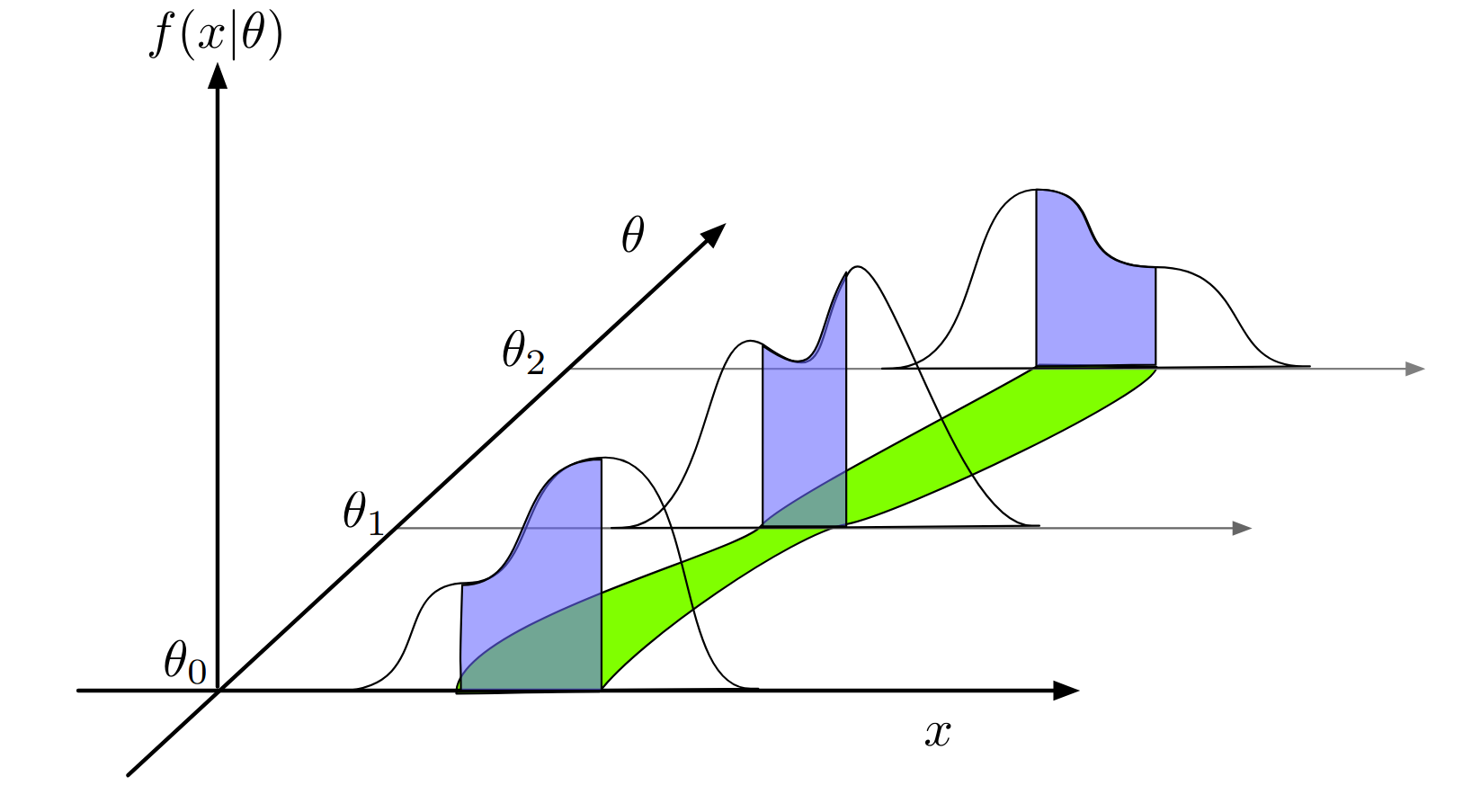
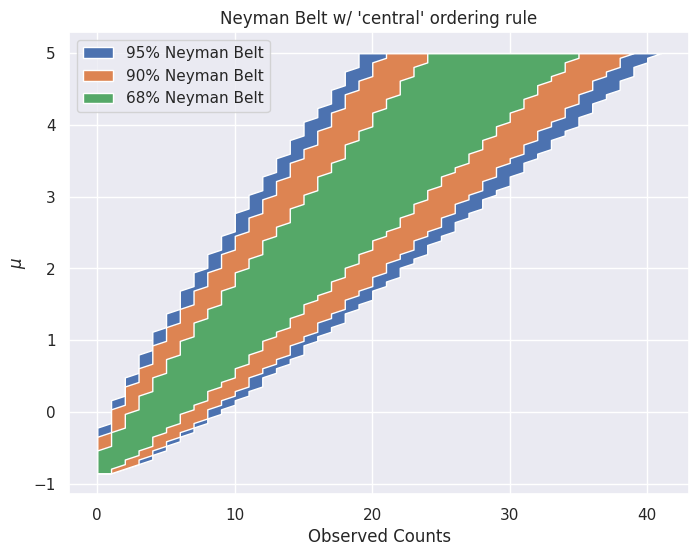
mu = 1
n_s = 5
n_b = 4.5
n_hypo = mu * n_s + n_b
CL_s = [0.68, 0.90, 0.95]
ordering_rules = ["central"]
n_plots = len(ordering_rules)
mus = np.linspace(-2, 5, 111)
fig, axs = plt.subplots( nrows=1, ncols=n_plots, figsize=(8 * n_plots, 6))
for i, ordering_rule in enumerate(ordering_rules):
ax = axs[i] if n_plots > 1 else axs
for CL in CL_s[::-1]:
n_hypo = mus * n_s + n_b
size = (1 - CL) / 2 if ordering_rule == "central" else (1 - CL)
lower_bound = poisson.ppf(size, n_hypo) if ordering_rule == "central" else np.zeros_like(n_hypo)
upper_bound = poisson.isf(size, n_hypo)
ax.fill_betweenx(mus, lower_bound, upper_bound,
label=f"{int(CL*100)}% Neyman Belt")
ax.set_title(f"Neyman Belt w/ '{ordering_rule}' ordering rule")
ax.set_xlabel("Observed Counts")
ax.set_ylabel(r"$\mu$")
ax.legend()
ax.grid(True)
plt.show()
Pyhf#
import pyhf
from pyhf.contrib.viz import brazil
import numpy.random as random
random.seed(0)
pyhf.set_backend("numpy")
model = pyhf.simplemodels.uncorrelated_background(
signal=[10.0], bkg=[50.0], bkg_uncertainty=[np.sqrt(50)]
)
observations = [60]
data = observations + model.config.auxdata
print(f" channels: {model.config.channels}")
print(f" nbins: {model.config.channel_nbins}")
print(f" samples: {model.config.samples}")
print(f" modifiers: {model.config.modifiers}")
print(f"parameters: {model.config.parameters}")
print(f" nauxdata: {model.config.nauxdata}")
print(f" auxdata: {model.config.auxdata}")
channels: ['singlechannel']
nbins: {'singlechannel': 1}
samples: ['background', 'signal']
modifiers: [('mu', 'normfactor'), ('uncorr_bkguncrt', 'shapesys')]
parameters: ['mu', 'uncorr_bkguncrt']
nauxdata: 1
auxdata: [np.float64(49.99999999999999)]
init_pars = model.config.suggested_init()
par_bounds = model.config.suggested_bounds()
fixed_params = model.config.suggested_fixed()
poi_values = np.linspace(0.1, 5, 50)
obs_limit, exp_limits, (scan, results) = pyhf.infer.intervals.upper_limits.upper_limit(
data, model, poi_values, level=0.05, return_results=True
)
print(f"Upper limit (obs): μ = {obs_limit:.4f}")
print(f"Upper limit (exp): μ = {exp_limits[2]:.4f}")
Upper limit (obs): μ = 2.8447
Upper limit (exp): μ = 2.0751
fig, ax = plt.subplots()
fig.set_size_inches(10.5, 7)
ax.set_title("Hypothesis Tests")
artists = brazil.plot_results(poi_values, results, ax=ax)
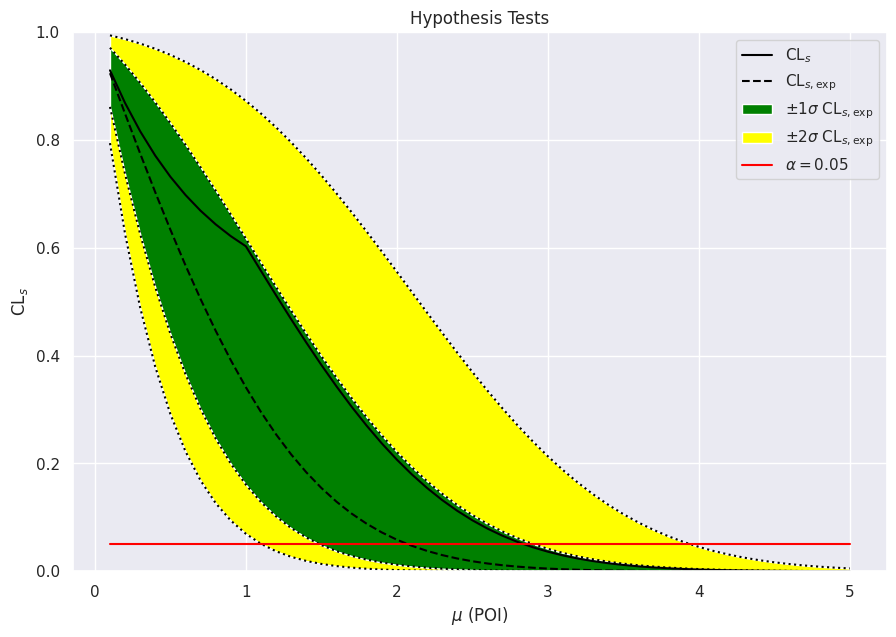
How is this related to the reported upper limits?
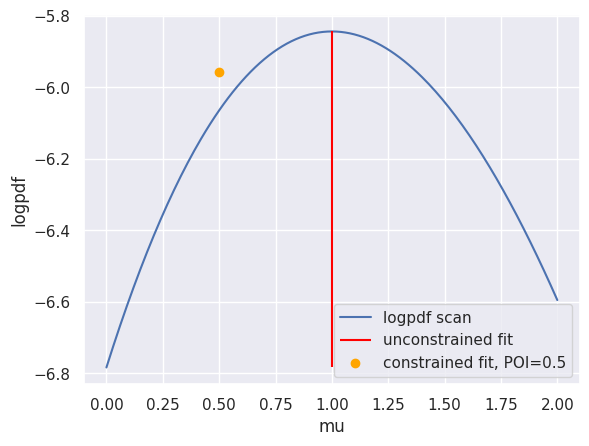
log pdf scan#
mu_s = np.linspace(0, 2, 201).reshape(201,1)
test_pars = np.concatenate((mu_s, np.ones_like(mu_s)), axis=1)
logpdf_scan = []
for test in test_pars:
logpdf_scan.append(model.logpdf(pars=test, data=data))
plt.plot(mu_s, logpdf_scan, label="logpdf scan")
uncontrained_fit, uncontrained_fitted_value = pyhf.infer.mle.fit(data=data, pdf=model, return_fitted_val=True)
constrained_fit, contrained_fitted_value = pyhf.infer.mle.fixed_poi_fit(poi_val=0.5, data=data, pdf=model, return_fitted_val=True)
plt.vlines(uncontrained_fit[0], ymin=np.min(logpdf_scan), ymax=np.max(logpdf_scan), label="unconstrained fit", color="red")
# plt.scatter(uncontrained_fit[0], uncontrained_fitted_value, label=f"unconstrained fit")
plt.scatter(constrained_fit[0], model.logpdf(pars=constrained_fit, data=data), label=f"constrained fit, POI={constrained_fit[0]}", color="orange")
plt.xlabel('mu')
plt.ylabel('logpdf')
plt.legend()
plt.show()
upper limits#
mu_test = 1.0
toy_calculator = pyhf.infer.calculators.ToyCalculator(
data, model, ntoys=2000, track_progress=False, test_stat="qtilde"
)
q_tilde = toy_calculator.teststatistic(2 * mu_test)
sig_plus_bkg_dist, bkg_dist = toy_calculator.distributions(mu_test)
bin_edges = np.linspace(0, 2, 21)
plt.hist(sig_plus_bkg_dist.samples, bins=bin_edges, label="s+b", alpha=0.5)
plt.hist(bkg_dist.samples, bins=bin_edges, label="bonly", alpha=0.5)
plt.vlines(q_tilde, 0, 1000, label=f"q_tilde={q_tilde:.2f}")
plt.legend()
plt.show()
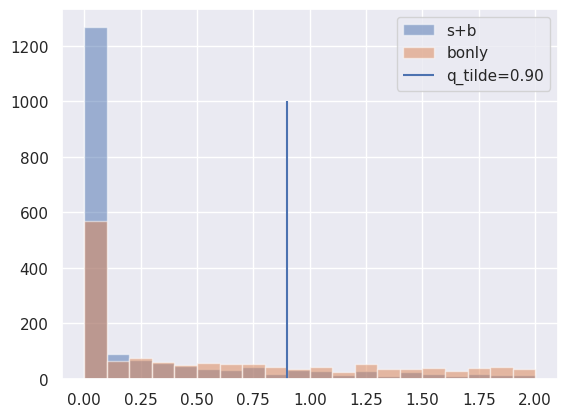
Observed Upper Limit#
CLsb, CLb, CLs = toy_calculator.pvalues(q_tilde, sig_plus_bkg_dist, bkg_dist)
CLsb, CLb, CLs
(array(0.1715), array(0.488), array(0.35143443))
print(sum(sig_plus_bkg_dist.samples > q_tilde) / len(sig_plus_bkg_dist.samples))
0.1715
sum(bkg_dist.samples > q_tilde) / len(bkg_dist.samples)
np.float64(0.488)
Expected Upper Limit#
CLsb_exp_band, CLb_exp_band, CLs_exp_band = toy_calculator.expected_pvalues(sig_plus_bkg_dist, bkg_dist)
CLsb_exp_band
[array(0.002), array(0.025), array(0.17775), array(1.), array(1.)]
CLb_exp_band
[array(0.0235), array(0.16), array(0.50125), array(1.), array(1.)]
[CLsb / CLb for CLsb, CLb in zip(CLsb_exp_band, CLb_exp_band)]
[np.float64(0.0851063829787234),
np.float64(0.15625),
np.float64(0.3546134663341646),
np.float64(1.0),
np.float64(1.0)]
CLs_exp_band
[array(0.10606061), array(0.16272189), array(0.35569106), array(1.), array(1.)]
test unc size#
import ipywidgets as widgets
@widgets.interact(bkg_unc=(1.0, 20.0, 0.5)) # bkg unc in [0, 10]
def test_unc_size(bkg_unc = 1.0):
model = pyhf.simplemodels.uncorrelated_background(
signal=[10.0], bkg=[50.0], bkg_uncertainty=[bkg_unc]
)
observations = [60.0] + model.config.auxdata
poi_values = np.linspace(0.1, 5, 50)
obs_limit, exp_limits, (scan, results) = pyhf.infer.intervals.upper_limits.upper_limit(
observations, model, poi_values, level=0.05, return_results=True
)
print(f"Upper limit (obs): μ = {obs_limit:.4f}")
print(f"Upper limit (exp): μ = {exp_limits[2]:.4f}")
fig, ax = plt.subplots()
fig.set_size_inches(10.5, 7)
ax.set_title("Hypothesis Tests")
artists = brazil.plot_results(poi_values, results, ax=ax)
